Polymerase chain reaction, or PCR, is a commonly used laboratory technique that was invented in the 1980s. The method has many applications in different fields, ranging from identifying individuals in forensic science, detecting pathogens in water supply, and genetic testing in medicine.
PCR works by first obtaining a sample that contains genetic information (DNA) such as blood. Then using a specialized enzyme called “DNA polymerase”, researchs can amplify and make billions of copies of a specific segment within the DNA that they are interested in (the region of interest). Because of this amplifying capacity, PCR is a very sensitive test and can be useful to detect even small trace amounts of DNA in a sample.

By producing billions of copies of DNA, it also makes it possible for scientists to analyze the region of interest. To amplify only the region of interest within the original DNA sample, scientists design and use “primers”. Primers are very short single-stranded DNA sequences specifically designed to match up with a specific region of interest within the DNA. Two primers are used because one is for the beginning of the sequence, and the other is for the end.
How is a PCR test done in the lab?
To perform the PCR reaction in the laboratory, scientists mix the genetic sample, the primers, some nucleotides (the building blocks of DNA), minerals (such as magnesium), and the important enzyme DNA polymerase in a small test tube. The test tube is put into a thermocycler, a device that can quickly and accurately change temperatures. In order for the DNA replication process proceed, there needs to be mutiple suddent changes in tempurature.
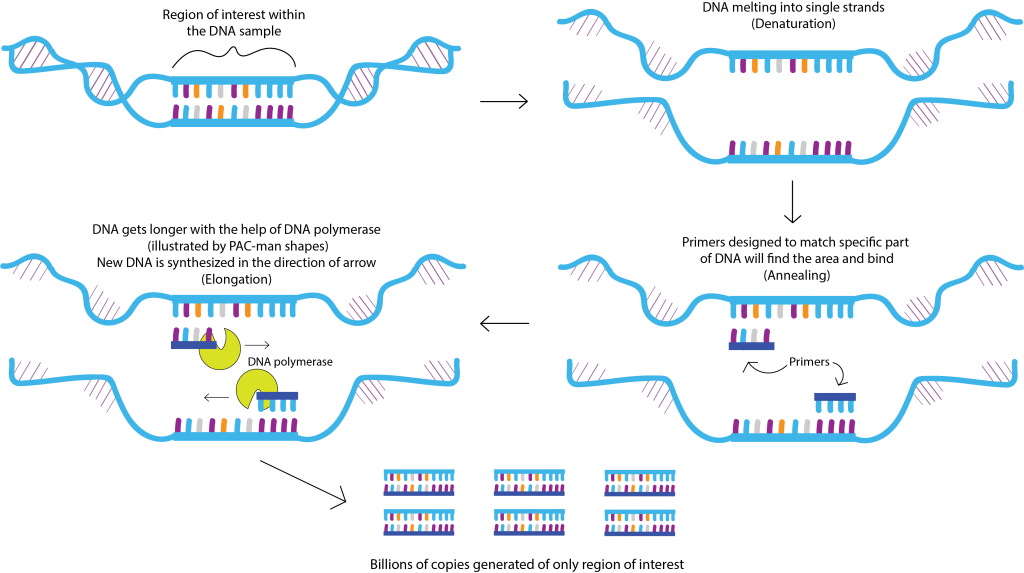
The first step of the cycle is called “denaturation”. This is where the genomic DNA from the sample is melted to a very high temperature of 96 °C. This makes the double-stranded DNA becomes single-stranded.
The next step is called “annealing”, where the temperature is reduced to 55 °C. This lets the primers can bind to the region of interest within the single-stranded genomic DNA.
Then, “elongation” occurs where the temperature is raised to 72 °C. This allows the DNA polymerase to effectively replicate the DNA bound by primers to synthesize double-stranded DNA by adding nucleotides. This process cycles from 20-40 times, and each cycle exponentially increases the quantity of DNA.
How is PCR used in ataxia research and treatment?
Certain types of ataxia have specific mutations to DNA that cause disease. For example, some types of spinocerebellar ataxia are caused by a type of mutation called polyglutamine expansion. The tests used to diagnose these types of ataxia use PCR to check to see if the mutations are there. Researchers also use PCR to confirm that cells, animals, or humans have the DNA mutation that researchers want to study. By checking this, researchers know what samples they are working with before starting their experiments.
If you would like to learn more about polymerase chain reaction, take a look at these resources by the Science Learning Hub and Your Geonome.
Snapshot written by Nola Begeja and edited by Larissa Nitschke










